Democratizing Genomic Research: nf-core and the Power of No-Code, Reproducible Pipelines
July 27, 2025
·Layla Bitar & Abdullah Atia (Co-author)
Let me confess something right away: I’m not a bioinformatician.
But as I started digging into the world of bioinformatics, one name kept popping up: nf-core.
And the more I learned about it, the more I realized this wasn’t just a technical tool for scientists in lab coats; it’s something that is quietly powering RNA sequencing, omics data analysis, and workflow automation in research labs around the world.
So, what is nf-core, and why should we care?
Before we dive into the science, let’s talk about food.
Imagine you’re making a lasagna. You could freestyle it, of course, toss in some noodles, sauce, cheese, and hope for the best. But if your friend across the country tries to do the same, chances are your results will look (and taste) pretty different.
Now, imagine there’s a single, perfected, step-by-step lasagna recipe that anyone can follow; and every time, you get a delicious, consistent dish.
That’s exactly what nf-core offers to scientists trying to “cook” complex analysis of genetic data.
If you're like me and new to this world, here's a super simple way to think about it:
Bioinformatics is what happens when we mix biology with data science. Scientists use software to analyze huge amounts of genetic information (DNA, RNA, proteins) to figure out what’s going on in a cell, a disease, or a patient.
But analyzing this data isn’t just a single step. It’s a pipeline: a series of connected tasks that clean, process, compare, and interpret the genetic code.
These tasks can get technical fast, often requiring multiple tools, and programming languages such as Python and R. And that’s where the magic (and frustration) often lies.
Here’s something I learned quickly: without standardization, bioinformatics can become a mess.
- Every lab might build its own pipeline.
- One person uses Tool A, someone else uses Tool B.
- Results might vary depending on the software version, the server, or even the operating system.
- Reproducing someone else’s analysis is often a nightmare.
It’s like everyone’s making lasagna with different recipes, and somehow you’re supposed to compare them and say which is best. Good luck with that.
Enter nf-core: The Master Cookbook
nf-core is a community-driven project that builds high-quality, ready-to-use pipelines for bioinformatics. Think of it as a carefully curated collection of trusted recipes; each one battle-tested, peer-reviewed, and open to anyone who wants to use or improve it.
Developed using a tool called Nextflow (which you can think of as a pipeline engine), nf-core pipelines are:
- Standardized: Everyone follows the same steps.
- Reproducible: You get the same result, every time, everywhere.
- Portable: Whether you’re on a laptop, a supercomputer, or the cloud — it works.
- Community-maintained: Scientists around the world contribute to improving them.
And the best part? You don’t need to be a hardcore coder to use them.
Let’s Paint a Picture: A Day in the Life of an nf-core Pipeline
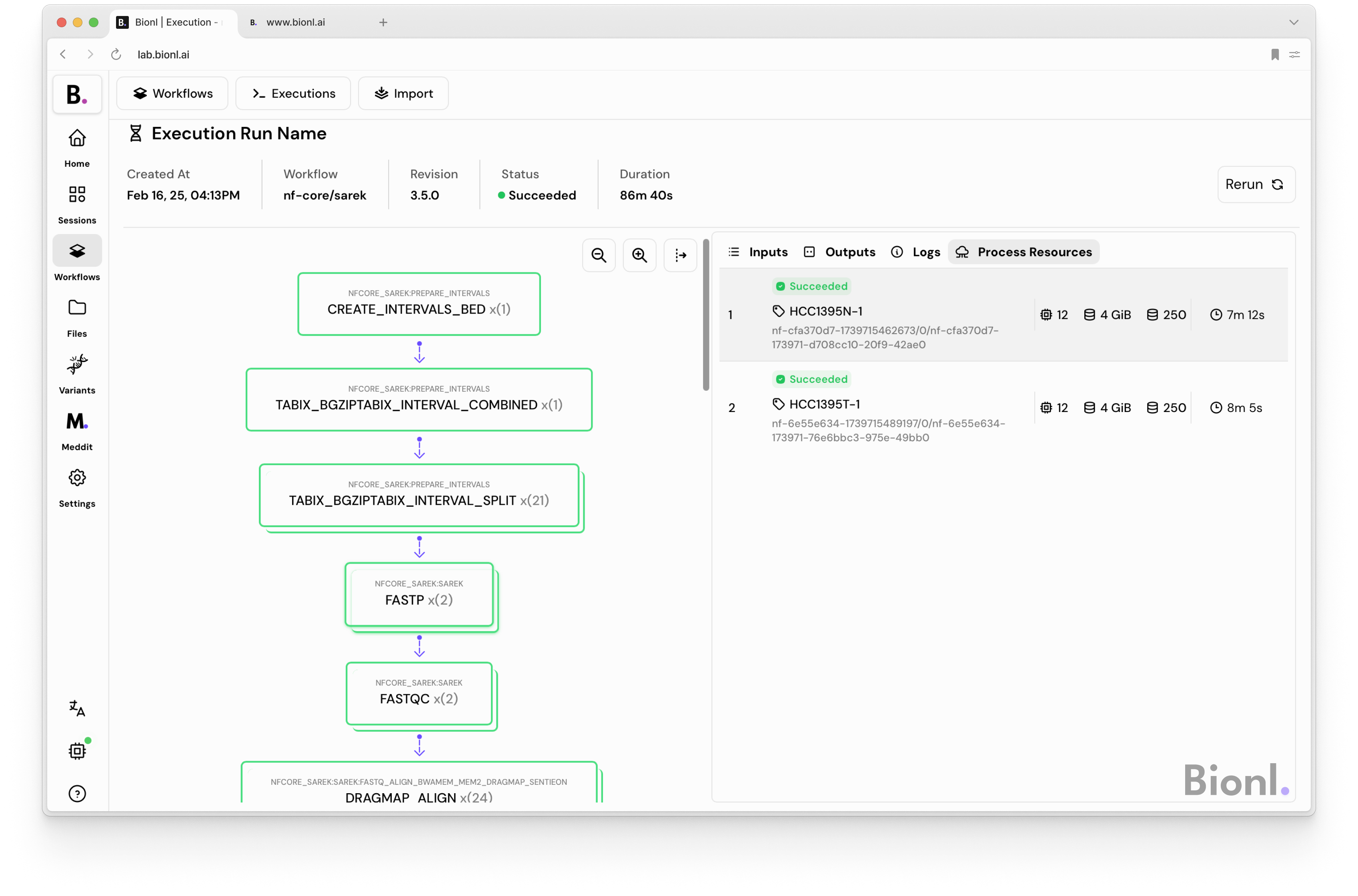
Imagine a hospital wants to analyze the DNA of cancer patients to look for mutations that could guide treatment. They receive raw sequencing data, basically, long strings of letters (A, T, C, G).
Without a tool like nf-core, this would require a team of bioinformatics experts and a lot of trial-and-error coding.
But with nf-core/sarek, one of the many ready-made pipelines, the hospital can:
- Clean and align the data
- Identify genetic variants
- Generate clear, detailed reports
- All in a standardized, reproducible, and fast manner.
That’s not just convenience; that’s life-saving insight, delivered faster.
Final Thoughts
As someone who came into this topic as an outsider, I now see nf-core not as a niche toolkit, but as a quiet powerhouse. It enables discoveries, streamlines workflows, and — perhaps most importantly — makes science more open, equitable, and reproducible.
But for someone with a limited coding background, I have resorted to Bionl, which has made nf-core pipelines much more accessible.
No command lines, no coding. Rather, we built a clean, intuitive interface where you can run complex genetic analyses in just a few clicks. It is a no-code platform with enabled cloud automation and workflow management.
Whether you’re a researcher, clinician, or simply curious about the future of genomics, Bionl lets you harness cutting-edge bioinformatics; no PhD required.
Explore Bionl and start managing workflows the easy way.
Related Posts
- Bioinformatics
- Genomic
Vaccines and Genomics: A Synergistic Approach to Disease Prevention
In the battle against infectious diseases, vaccines have been our most potent weapon. The advent of genomics and bioinformatics has revolutionized vaccine development, enabling more precise and rapid responses to emerging threats. This blog explores the relationship between vaccines and genomics, the importance of understanding genomic data in vaccine discovery, and how bioinformatics might accelerate the process of vaccine discovery.
Aug 2023
Tasnim Nour
- Literature Search
- Artificial Intelligence
Helpful or Harmful? How to Trust AI-Generated Summaries
AI summaries can support research and clinical work in several complementary ways. They can condense long and complex text into clear takeaways such as the primary endpoint, population size, and outcomes, giving readers a head start before committing to the full text.
Oct 2025